
Prof. Chunhai FAN, academician of Chinese Academy of Science, and Prof. Fei WANG from the School of Chemistry and Chemical Engineering and the Frontiers Science Center for Transformative Molecules have recently pioneered the development of a DNA-based programmable gate array (DPGA). DPGA enables universal digital DNA computing through molecular implementation, facilitating the construction of large-scale, liquid-phase molecular circuits with minimal signal decay. This work has been published on the top journal Nature.
In 1994, Adleman, Turing Award laureate, proposed the concept of biomolecular computation based on the principle of base-pairing. Since then, liquid-phase DNA circuitry has held the potential for massive parallelism in both encoding and execution of algorithms. To date, computational DNA reaction networks including automata, logic circuits, decision-making machines and neural networks have been realized. These achievements have showcased potential applications in molecular information processing, synthetic intelligent devices, and biomedical applications. Despite this progress, most of these computing systems are tailored in hardware to implement a specific algorithm or a limited number of computational tasks. General-purpose electronic integrated circuits allow software programming rather than application-specific custom hardware fabrication to perform a certain function, providing a higher-level platform for prototyping computational machines without the requirement of previous knowledge of the underlying physics. Therefore, developing general-purpose computing and exploring its programmability and scalability has become a bottleneck in the field of DNA computing. This entails finding methods to achieve universality in DNA computing for more flexible execution of various computational tasks. DPGA is an attempt to progress towards this goal, making DNA computing more programmable and scalable.
To address the challenge, the authors first demonstrated that using generic single-stranded DNA oligonucleotides as a uniform transmission signal (DNA-UTS) can achieve functions like electronic signal transmission in circuits. Subsequently, they developed one DPGA that supports universal digital computing and enables multi-DPGA integration at the device level, thus achieving programmability within and among the devices. As is shown in Fig.1, when the complexity of a circuit exceeds the capability of a single DPGA, DPGA can be divided into subunits, for which molecular instructions were generated. Involved computing units of each subcircuit were called by their logical address and the function was implemented via intra-DPGA routing with molecular instructions. The transmission of intermediate output between subcircuits was realized via DNA origami register-mediated inter-DPGA routing.
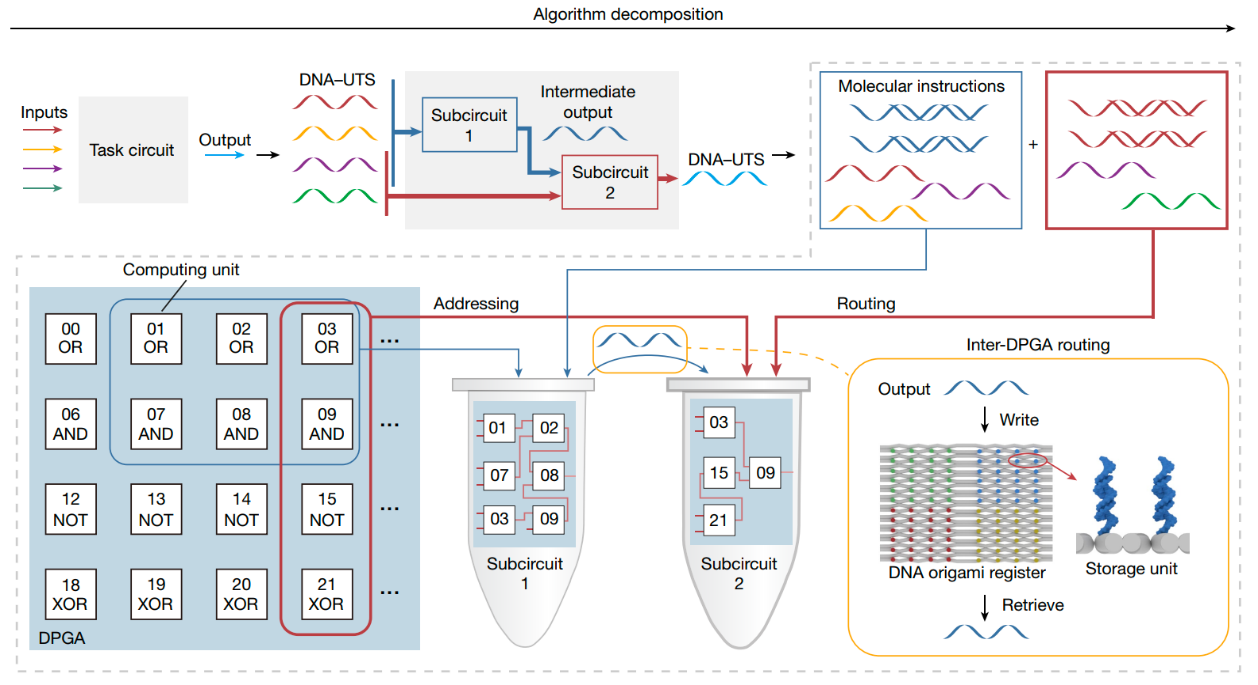
Fig.1: Schematic workflow of DPGA programming.
By leveraging the programmability and large-scale integration of DPGA, this work has overcome the limitations of circuit scale and depth in DNA computing. For the first time in experiments, it has demonstrated up to 30 logic gates, 500 participating strands, and 30 steps of cascaded strand displacement reactions (SDRs), which represents a significant breakthrough in the field of DNA computing in nearly two decades (Fig.2).
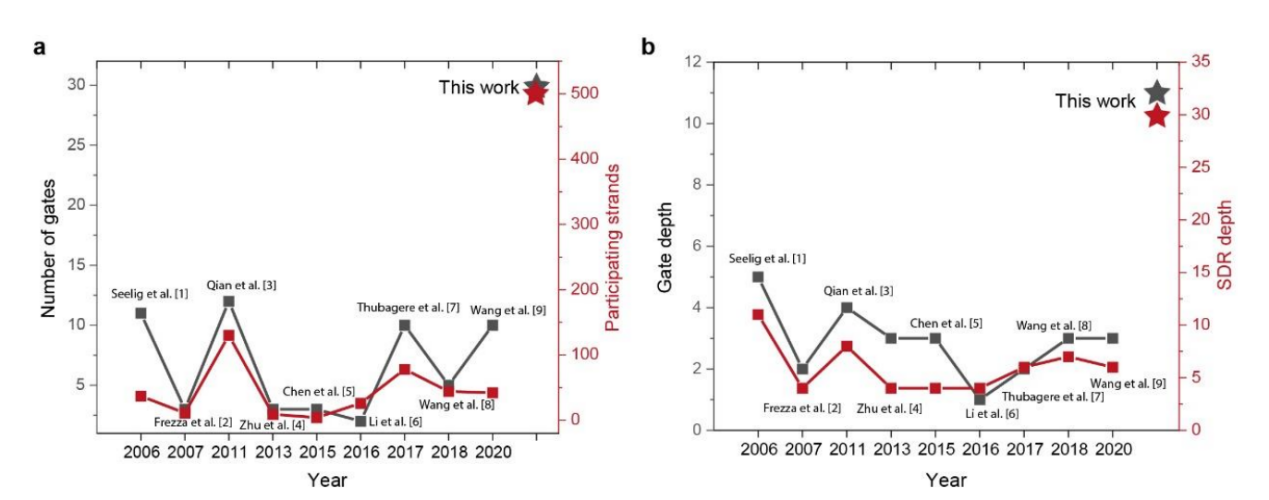
Fig.2: DPGA-based digital computing in comparison with existing SDR-based logic computing systems.
Further studies found no significant signal decay of DNA-UTS during transmission across multiple DPGAs, demonstrating the high scalability of DPGAs. This predicts that any practical problem can theoretically be plugged into a DPGA circuit via DNA-UTS after analog-to-digital conversion. The use of DPGA as an information processing core in molecular diagnostics for nonlinear classification of disease-related molecular targets is conceptually demonstrated in this study.
This work was supported by the National Key R&D Program of China (grant no. 2021YFF1200300), the National Natural Science Foundation of China (grant nos. T2188102, 21991134, 21904060, 22025404 and 22104088), the Science Foundation of Shanghai Municipal Science and Technology Commission (grant nos. 20dz1101000 and 21TQ1400222) and the New Cornerstone Investigator Program.
Link to article: https://www.nature.com/articles/s41586-023-06484-9
Translator: Chenyun SUN
Reviser: Xiaoke HU
Address:800 Dongchuan RD. Minhang District, Shanghai, China
PostCode:200240 Tel:021-54742893 E-mail:sjtuscce@sjtu.edu.cn

Copyright@ 2023. All rights reserved. Powered by SCCE ICP:2010917